Current Research Directions
- Functions of potassium channels and bioelectricity in vertebrate development and tumorigenesis.
- Cancer driver gene discovery on the aneuploid chromosomes: Comparative cancer genomics and phylooncogenomics
- Cancer genetics: The zebrafish model of human diseases
Functions of potassium channels and bioelectricity in vertebrate development and tumorigenesis.
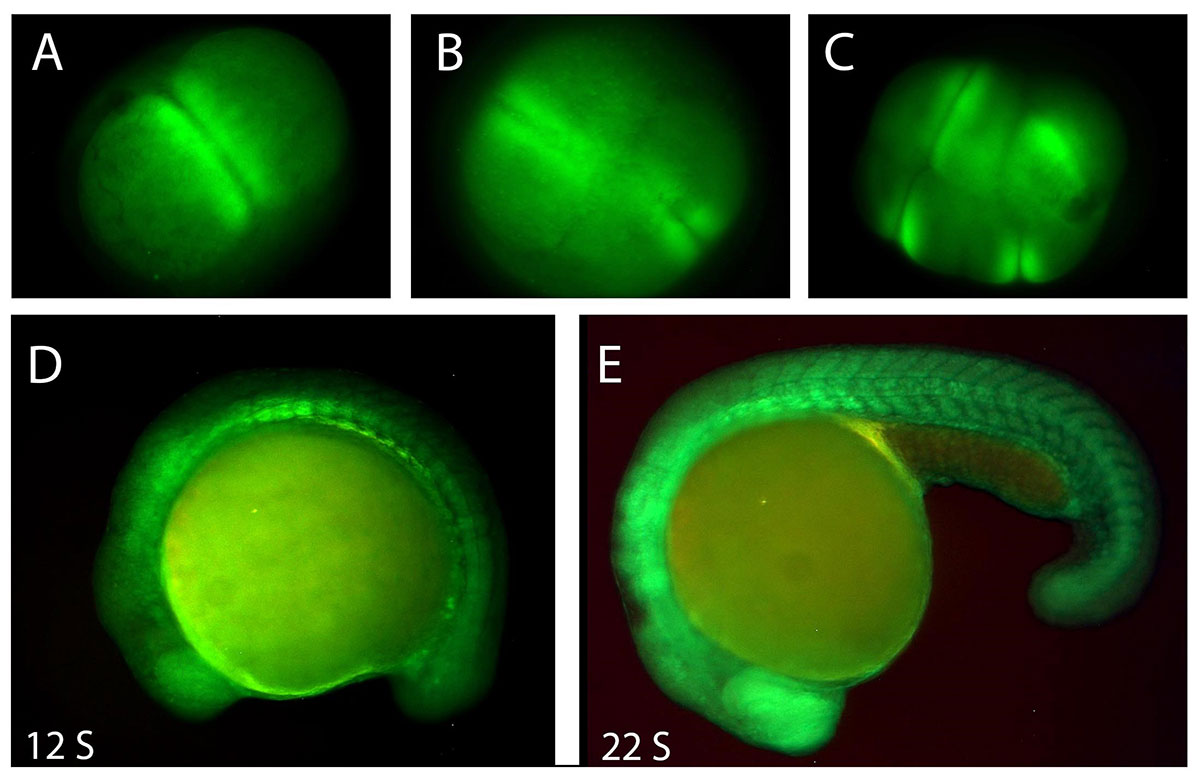
Potassium channels and their related bioelectricity have long been known for their roles in regulating neuronal excitability, neurotransmitter release, and muscle contraction. Mutations or malfunctions of potassium channel genes cause a variety of diseases in the central nervous system, heart, pancreas, and kidneys (long-QT syndromes, episodic ataxia, familial convulsions, etc.). There is growing evidence that bioelectricity also plays an important role in vertebrate embryogenesis, wound healing, and cancer.
Bioelectricity refers to endogenous electrical signaling mediated by ion channels and pumps located throughout the cell membrane. Cell membranes act as electrical insulators, in part, due to the phospholipid bilayer’s impermeability to ions. Any ion that forms a gradient across a membrane will contribute to the actual membrane potential and bioelectrical signaling.
Currently, we are focusing on the developmental roles of potassium channel genes zebrafish embryos and their oncogenic functions in adult zebrafish.
Selected Publications
Silic M.R., Zhang G. (2018). Visualization of Cellular Electrical Activity in Zebrafish Early Embryos and Tumors. JOVE , 134: e57330, doi:10.3791/57330
Cancer driver gene discovery on the aneuploid chromosomes: Comparative cancer genomics and phylooncogenomics
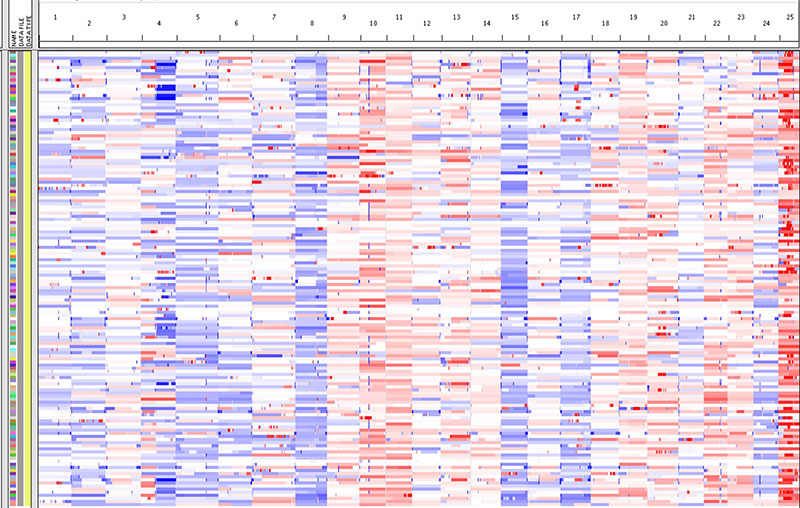
Cancer is currently known as a genetic or genomic disease. During the development of cancer, many mutations are accumulated in the cancer cell genomes. Aneuploidy, the numeric alterations of chromosomes, is one of the most common genetic changes in cancer cell genomes. As chromosomes carry many genes, this aneuploidy influences their gene expression through gene dosage adjustment. Similar with other types of genetic mutations, not all the genes on the aneuploid chromosomes equally contribute to cancer formation. The critical genes that contribute to tumorigenesis are called “driver genes”, while genes that do not contribute are called “passenger genes”. One of the goals of current cancer research is to identify cancer driver genes, so we can use them as therapeutic targets as well as diagnosis and prognosis makers.
The next generation sequencing, aka, massively parallel DNA sequencing, is newly developed powerful technology, which is very effective for finding point mutations on a single nucleic acid level. However, next generation sequencing looses its power with aneuploidy chromosomes, because these chromosomes were always found as large genetic changes and the technology can not distinguish drivers since there are many genes are resided on the same chromosome. To overcome this difficulty, we are taking the approach of comparative cancer genomics for finding out the cancer driver genes on the aneuploid chromosomes. The comparative cancer genomics is based on the syntenic relationships among different vertebrate species. In vertebrates, the genes’ functions are usually conserved, while their physical position on the chromosomes are not fixed on specific chromosomes. From the evolutionary point of view, cancer is a process of somatic evolution, which is similar with species evolution. During this process, driver genes are selected for gains and losses depend on the needs of cancer cell survival. Thus, cancer driver genes can be enriched by comparing the DNA copy number alterations (CNAs) from cancer genomes of two vertebrate species. We have found zebrafish-human cancer genome comparison is more effective than mammalian species because zebrafish and human share small sytenies. Phylooncogenomics could be achieved in the future by introducing more different species.
The long-term goal of this research direction is to better understand the nature of cancer by putting cancer into a vertebrate evolutionary context.
Selected Publications
Zhang G., Hoersch S., Amsterdam A., Whittaker C.A., Beert E., Catchen, J.M., Farrington, S., Postlethwait, J.H., Legius E., Hopkins, N., Lees, J.A. (2013) Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery. PLoS Genetics 9(8): e1003734. doi:10.1371/journal.pgen.1003734.
Zhang G., Vemulapalli, T.H., Yang J.Y. (2013) Phylooncogenomics: Examining the cancer genome in the context of vertebrate evolution. Applied & Translational Genomics , 2: 48-54.
Cancer genetics: The zebrafish model of human diseases

Zebrafish is one of the prominent vertebrate model organisms. It is widely used in the field of developmental biology due to its large number of progeny, early transparent embryos, and tractable genetics and ease in maintaining. Recently, zebrafish is also found their way into other research areas including cancer research. It has been found that gene mutations of many human cancer gene homologues also lead to cancers in zebrafish. And zebrafish and human share similar characters of cancer genomics and transcriptomes, such as aneuploid chromosome and chromosome translocations. Currently, we are focusing on the flowing directions:
1. Identifying novel tumor suppressors on chromosome 9p, which is one of the frequently lost chromosome arms in human solid tumors including carcinomas and sarcomas. Using zebrafish-human comparative oncogenomic analysis, we have identified 36 candidate genes (KANK1, RECK and others). We have made zebrafish mutants on some of the candidate genes using CRISPR, and we are currently assessing these candidate genes’ functions in both zebrafish and human cancer cell lines.
2. Identifying novel oncogene candidates on human chromosome 4q12. In cancer cell genomes, focal CNAs are generally not as common as than the large ones (i.e. aneuploidy). In one of our past studies, we only found one focal CNAs (4q12) shared by zebrafish and human malignant peripheral sheath tumors (MPNST). This suggested that there are important cancer drivers genes within this region. Indeed, c-Met, PDGFR and MDM2 are located in this region. While other genes in this region largely remain unexplored. One of the candidates is MDM1, which is amplified in many human cancers according to the cancer genome database. So far, we have made zebrafish loss-of-function mutants and transgenic lines on this gene to reveal its functions in cancers.
3. Zebrafish model of human neurofibromatosis Type II (NF2). NF II is a relatively common hereditary human disease with an approximate occurrence of 1 in 40,000 in human population. It is generally caused by the loss of function of NF2 gene. The typical symptom of the neurofibromatosis type II is bilateral vestibular schwannomas, which occurs on the 8th cranial nerve. Except for the bilateral vestibular schwannomas, the patients usually have other neural tumors (meningioma, glioma etc), neuropathies (hearing loss, disequilibrium, facial numbness and weakness) and cataract. Moreover, recent studies revealed that NF2 gene mutations are common in many other types of human cancers and that NF2 plays important roles for normal neural development. In zebrafish, there are two NF2 homologous genes, nf2a and nf2b. Nf2b was found to be a tumor suppressor in zebrafish, while the function of nf2a remains unknown. We have created nf2a mutants using CRISPR technology, and are currently in the process of elucidating the detailed molecular and cellular tumorigenic mechanisms and roles of nf2 gene during zebrafish development.
Selected Publications
Zhang G., Hoersch S., Amsterdam A., Whittaker C.A., Beert E., Catchen, J.M., Farrington, S., Postlethwait, J.H., Legius E., Hopkins, N., Lees, J.A. (2013) Comparative Oncogenomic Analysis of Copy Number Alterations in Human and Zebrafish Tumors Enables Cancer Driver Discovery. PLoS Genetics 9(8): e1003734. doi:10.1371/journal.pgen.1003734.
Zhang G., Hoersch S., Amsterdam A., Whittaker C.A,. Lees J.A., Hopkins N. (2010). Highly aneuploid zebrafish malignant peripheral nerve sheath tumors have genetic alterations similar to human cancers. Proceedings of the National Academy of Sciences , 107: 16940–16945.

